SBW for a long time had facilities for exporting SBML models to Matlab and a slew of other formats. I have to admit I never really used the Matlab Export before, and so it came as a surprise when I heard that the export of SBML Assignment rules was broken. But good news, at least this export is now working just fine.

Here we set Node0 and Node2 to be a Boundary (fixed) Species, as they are not really part of the system. And we want Node0 to be controlled by an Assignment Rule, which sets the value of Node0 to ‘5’ starting at 1 second for 2 seconds. Outside this window Node0 should be close to zero (and I say close to zero and not zero so that the integrators will pick up the change). So this translates into a piecewise function:
Node0 = 5 if 1 < time < 3 and 0.1 otherwise
in SBML infix speak this translates into:
Node0 = piecewise(5, and(geq(time,1), leq(time, 3)), 0.1)
here of course piecewise, stands for the piecewise operation, note that you can have as many pieces as you’d want … for every piece you would add two terms, the value and a Boolean expression. The Boolean expressions supported are:
| Name | Argument type | Explanation |
| and | n-ary | n-ary and like in the example above, it is 1iff all arguments are 1 |
| xor | n-ary | n-ary xor, is 1 iff not all arguments are 1 or all arguments are 0 |
| or | n-ary | n-ary or, is 1 if at least one of the arguments is 1 |
| not | unary | is 1 if argument is 0 and 0 otherwise |
| gt(a,b) | binary | greater: is 1 if a > b |
| lt(a,b) | binary | less than: is 1 if a < b |
| geq(a,b) | binary | greater or equal: is 1 if a >= b |
| leq(a,b) | binary | less or equal: is 1 if a <= b |
| eq(a,b) | binary | equal: is 1 iff a == b |
| neq(a,b) | binary | not equal: is 1 iff a != b |
| piecewise( value, condition, otheriwse) | n-ary | piecewise function: each piece is defined with a value, condition pair and concluded with an value for the otherwise clause. all values and conditions can of course be function expressions as well. |
So let us look at the complete model in the JarnacLite language (this will allow you to just paste it into JarnacLite without you having to download the SBML model):
p = defn AsssignmentExport
var Node1;
ext Node0,Node2;J0: Node0 -> Node1; J0_k*Node0;
J1: Node1 -> Node2; J1_k*Node1;Node0 = piecewise(5, and(geq(time,1), leq(time, 3)), 0.1);
end;
p.Node0 = 0;
p.Node1 = 0;
p.Node2 = 0;
p.J0_k = 0.1;
p.J1_k = 0.1;
When in JarnacLite, a click on SBW\Translate SBML –> Any will bring up the SBML Translators, and a click on the Matlab tab shows you the newly translated model:
As you can see this model is complete with instructions on how to use it in Matlab. And here the good news it works in Octave as well! (you’d just download ode23.m and put it in the same directory and you should be good to go!) So all that is left is to save the file as .. say … AsssignmentExport.m into your Matlab work directory and run:
x0 = AsssignmentExport
[t, x] = ode23s(@AssignmentExport, [0 20], AssignmentExport);
plot(t,x);
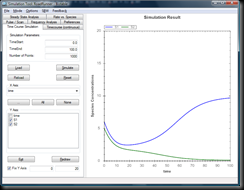
and you should see a graph like this:
And here the disclaimer, I do know that you’d normally implement these sort of things as SBML Events, but so far we don’t export SBML events to Matlab, but you can always use roadRunner to have them simulated correctly.
So if you want to give this new version of the Matlab export a try, download a new Installer, or select Systems Biology Workbench\Utilities\Update SBW from an existing SBW installation.