I was asked to give a brief talk for the Semantic Web in Biomedicine Seminar, on computational tools that make the task of using all the great ontologies (like MIRIAM, SBO, ChEBI, UniProt, KEGG) that we have come to love easier.
I will add the full presentation below. However, as always without all too much text, thus I’m going to continue below.
I’m deeply involved with the Systems Biology Workbench (SBW), a lightweight framework that enables applications to share their functionality regardless of operating system or programming language. With SBW we also deliver a full toolset for modeling, simulating and analyzing SBML models. About five years back we started to get involved with SBGN. So today it would be great if we could automatically generate SBGN diagrams out of our SBML models. What is needed for that are annotations. Not just any annotations but annotations in the form of SBO ( the material entity branch) would be perfect. Unfortunately this is not quite enough. Additional MIRIAM annotations are needed for example to describe complexes in more detail.
I’ve been looking at this for some time and there are a couple of interesting developments to mention, of course first and foremost the EBI Web Services. They certainly make the work much easier. I introduce the:
- BioModels Web Service
- SBO Web Service
- MIRIAM Web Service
- Ontology Lookup Web Service
Each web service is briefly tested, using a Generic Soap Client, as this will allow to show the sort of information that is being provided first hand.
Of course it would be great if one would not have to hunt each Web Service down and it would seem that others had the same idea. I mention libAnnotationSBML first, a neat Java library that promises to provide a unified interface for a whole list of web services:
(photo grabbed from Neil’s presentation on Slideshare). The only drawback I could find with it is that the library is IMHO not quite ready to use by non Java geniuses. Everyone hearing me: If you are developing a library … especially a library for Java / .NET or any of the other cross platform virtual machines … please please provide a binary for people to use. It’s been a while that I worked with Java, and an ant build script that won’t work is trouble for me. As it turned out the build failed because just of a couple of missing jar files and a test failure due to changes in the SBO. Still at the end of it all I ended up with a jar file and no instructions on what to do with it. I’ve seen great demos of libAnnoationSBML with the SBMLReactionBalancer for example. This could be made available online as binary as is …
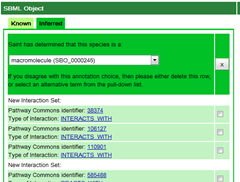
The presentation will go on to introduce SAINT: a lightweight integration environment for model annotation. It follows the same great idea as in libAnnoationSBML of hunting down the individual web services for annotations of your SBML file. However, whereas libAnnotationSBML is a Java library, SAINT is nifty web application based on the Google Web Toolkit(GWT). As such it really brings a long a snappy AJAX UI. SAINT has a lot of potential, but again it would seem I was out of luck, I could not manage to get a model annotated. Here is what went wrong: a) no upload button, but that is fine copying the SBML to clipboard and pasting it was not all that hard b) once I clicked “Annotate” however a whole bunch of Asynchronous worker bee's must have been sent out in order to fetch all my new Annotations. But when they came back they brought along a whole list of annotations I had no interested in:
The above image represents all the results found for “glucose”. And while it is great to find so many Pathway Commons Identifiers, the link to Pathway Commons lead to 404. The trick seems to be for me to disable the “New Reactions” checkbox in advanced options. Then I received a couple of SBO terms for my model, however not much more. But SBO terms are a good start, so from here I went to the “Get Annotated Model Code” tab, but try as I might I was never able to actually retrieve my annotated model.
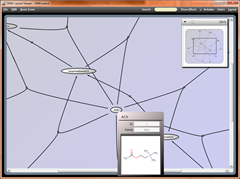
Looking at other currently available software tools like CellDesigner and COPASI, both tools that support MIRIAM annotations I am not sure whether users of the tools will take full advantage of the Annotation process. Thus I end the presentation with a couple of screenshots of software tools using the EBI Web Services, to interact with the BioModels Database, resolve the publication citation and visualize the annotations in a way better suited to the community.
Why should we just display the ChEBI identifiers, when we could just as well show a formula, or the name, with a click revealing the full set of annotations.


3 comments:
Hi Frank,
Thanks for the heads up. A response is below...
Computational Tools for the Annotation of SBML Models: the sequel
Cheers,
Neil.
I enjoyed over read your blog post. Your blog have nice information,
I got good ideas from this amazing blog.
goldenslot
gclub
gclub casino
Thank you for posting such a great article! I found your website perfect essay writing service of needs. It contains wonderful and helpful posts. Keep up the good work. Thank you for this wonderful. Also check Fotnight Gift Card
Post a Comment